New digital challenges in life science: the Swiss Lemanic area and beyond

The life science sector faces great challenges when the time comes to meet modern digital opportunities. This sector is incredibly dynamic, both from an economic and a scientific point of view, but innovation in the lab often comes in pair with a deep evolution in the information system strategy.
In this article, we will first focus on the Swiss Lemanic area landscape and see how the main computational trends encountered there are representative of the domain at large. We will then try to extract the most prevalent technical issues, either methodological or technological.
More importantly, we will see that life science has a particular culture and faces original computational issues. But a lot of the digital aspects are strikingly similar to those of other industries, like finance, retail or social media. Therefore, a great deal is to be learned from how companies in those sectors have undertaken their own transition towards a new digital era (and vice versa).
Welcome in Health Valley
The life science ecosystem consists of many actors: the pharmaceutical industry, biotechs, medical instrumentation companies and institutional centers. In Switzerland, this sector is dynamic, both scientifically and economically, and plays a major role in the country's economy [1] [2].
If Basel is the historical center and still remains the most active area, the Lemanic bassin (aka "the Health Valley") has no reasons to be shy. Science journal presumably even called the area "the number One cluster for life sciences research in continental Europe" [3].
Beside many private players (cf. figure 1), the public sector is also a key driver. Several remarkable public/private initiatives have been launched and provide a fertile breeding ground: Geneva Campus Biotech [4], EPFL Innovation Park [5], Lausanne Biopôle [6] etc.
We are right in the middle of it!
[caption id="attachment_56454" align="aligncenter" width="400"] 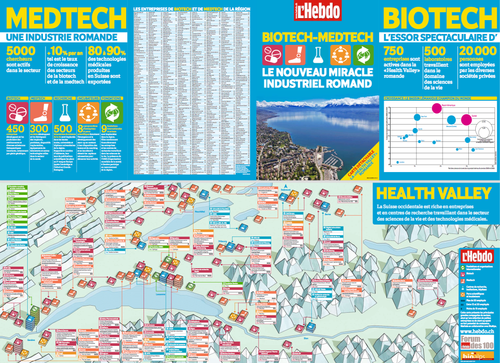
A 2010 panorama, but still relevant [7].
[/caption]
Where is the domain heading to?
It is far beyond this article's scope to depict the long term goals of all the private actors. Let's remain modest, and focus on the information system side of the challenges at stake. A few initiatives can be seen as representative:
- Health2020 [8]: the federal level comprehensive strategy, decided in 2013. Two of its highest priorities raise serious computational challenges: the electronic patient dossier and the handling of gigantic flows of personalized data.
- The Human Brain Project [9]: involving more than 130 partners in 26 countries, the project vision is to build a low level model of the human brain and to structure all the information on this subject. European by nature, the project is directed by EPFL, and its largest center is hosted in Geneva.
- CHUV Biobank [10]: one of the Lausanne University Hospital Biobank's mission is to collect patient data, like genomic sequences, at a very large scale. All consenting patients entering the hospital are targeted, with an estimation of 15,000 samples per year [11]. This project opens the path for a larger regional and country wide strategy.
These initiatives certainly shoot for the sky. But rather than focusing on technical (or scientific, ethical, political or even psychological) concerns raised by each of them, we should see these projects as representative of the stimulating local ambiance in life science and appreciate how they shape the Lemanic area landscape, both inwards and outwards.
Information System challenges
Addressing the core biological questions is also out of the scope of this paper and we'll focus on information system issues. Like other experimental sciences, the digital age has fully entered the wet lab. And some numerical challenges raise important, yet exciting questions.
Big Data
Let's shoot the elephant in the room first: laboratory instruments generate a huge information flow. Genomic sequencing is not the only data provider, but it is nowadays the most prominent one. The advent, a decade ago, of the Next Generation Sequencing (NGS) technology dramatically changed the paradigm. From three billions dollars in the nineties down to less than a thousand dollars today for a single individual [12], sequencing is making its way to the modern physician's toolbox.
[caption id="attachment_56453" align="aligncenter" width="400"]!["The $1,000 genome", Nature 12
"The $1,000 genome", Nature [12].
[/caption]Genomic sequencing covers a large variety of methods. A typical whole genome analysis, on a single patient sample, will generate roughly 200GB of data. And if we go back to the previous section, where sequencing 15,000 patients per year in a single hospital is the plan, we can estimate the total yearly volume to be around three petabytes (3x1015 bytes). The term "big data" gets all its weight.
As discussed in the aforementioned Nature paper, NGS broke the Moore's Law pattern. Keeping pace only with the data storage aspect of it is a tremendous challenge (not to mention analysis). On the one hand, compression efforts are undertaken to reduce data volume but hardly change the problem due to the exponential growth of said data. On the other hand, some scientists advocate to simply destroy the raw data and only keep some analyzed projections while storing the original samples in a freezer. They argue that freezer storage is much cheaper than disk space and if the raw sequence data are needed in the future, they could simply be acquired again (with certainly more performant technologies). Those ideas are still debated in the field as they face all sorts of ethical, scientific and technical arguments.
Visualization
Once terabytes have flowed to your hard disks and brilliant analysis methods have run on your large scale cluster, the problem is not over. In fact, it starts there: you have to try to make some sense out of this deluge of data. Static analysis reports are useful in some situations, but scientists often need to have a more intimate relationship with their data to question them, to navigate through them or to put them in perspective with other contexts.
Visualization, and more precisely reactive and interactive analysis, comes into action. Without requiring every tool to be as sophisticated as the Allen Institute Brain Atlas [13], interacting with data is often the best way to get a feel for and understand the underlying complexity.
It is to be noted that visualization can also encompass the "scientific art" category, producing striking (and beautiful) images [14]. But beside making a nice journal cover, or the best decorated office wall ever, such representations rarely convey deep and relevant scientific meaning.
Fortunately, the advent of powerful web browser technology, high speed network and strong cross domain emulation, paves the road to new possibilities. Publications and conferences do already exist in this field [15], but a new era is beginning.
The electronic patient dossier
Its goal is to digitize as much patient's data as possible, at a canton scale, and make them available to health sector actors (physicians, hospitals etc.) while hiding them to others (insurance companies, hackers). This goal stands high among the swiss Health2020 priorities. Without diving into the nitty-gritty details of such an endeavor, one can imagine the complexity of the underlying information system architecture.
Until now, only Geneva and Valais cantons have migrated to the electronic patient dossier. In fact, Valais almost made it, as the service was stalled after 4 days. To make a long story short, on August 27th, after four millions swiss francs of investment, the project was released. The communication department streamed messages about how secure the system would be, as "it had been heavily audited and proven unbeatable" [16]. So long for the unbeatable security: within a couple of days, the Pirate Party pinpointed "serious security concerns", waived out by freely available tools [17]. On August 31st, the Valais Minister of Health suspended the project [18], and everyone is now back to work...
[caption id="attachment_56452" align="aligncenter" width="400"] 
© Fran, cartoonstock.com
[/caption]
Biomedical sensors
Stepping across the three previous issues, this topic embraces the gathering, storage and analysis of physiological measures. Many devices allow for real time monitoring of heart rate, activity etc. both for medical and comfort reasons. This subject, sometimes referred to as "Quantified Self" [19], ranks high in the news and many actors eagerly look in that direction, although it is hard to predict its real importance on the longer run.
Laboratory Information Management System (and other heterogeneous data management)
A LIMS tracks all the laboratory operations on a biological sample. Where does it come from? How was it preprocessed? Which instrument did it go through? And which analysis steps were performed? What (and where) are the analysis results? This information is crucial for proof, audit, reproducibility, etc. and an efficient LIMS platform is a Grail for most laboratories.
If such platforms are not a new need, their actual implementations raise recurrent problems in the lab. Reality is often, at best, an assembly of custom and third party software. In fine, the scientist often falls back on his paper lab notebook as the single source of truth.
Technical background and issues in life science information systems
Stepping back from particular domain silos, we discussed how life science actors are facing recurring issues from an information system perspective. Let's now try to identify some of the reasons underlying those patterns and the different dimensions in which they occur. We'll finally realize how similar those aspects are to other domains and how lessons learned in other industries could be applied in life science.
Software development culture in life science
Let's consider the software development culture as the overall environment in which digital projects are built. It encompasses low level aspects, like programming languages and tools used by developers, but also processes, relationships with customers (aka the wet lab scientists) and within the development teams. Last but not least, it also covers how upper management envisions the information system at the organization scale.
Why is life science so special?
Because dry geeks (the ones with a strange look and a keyboard) have to talk to wet geeks (the ones with a strange look and a white coat). And also because trying to understand biological phenomena can be incredibly complex and always drives people into unforeseeable journeys.
Because of its culture, the life science digital ecosystem is original. A key factor is the porosity between academic and private companies and the historical importance of one's scientific knowledge to be recognized by his peers. This leads to situations where a large proportion of software development players, at all levels, are former biology or chemistry trained people (with a strong inclination towards a keyboard, indeed).
When the time comes to selecting a new hire, the balance often tips towards a biology fluent person with IT knowledge rather than towards a hard core software engineer, architect or manager, with proven abilities to dialog with people outside of her domain. Some companies try to counter that trend, but unfortunately "mirror hiring" is a natural tendency for most of us [ [20].
Technical bottlenecks
Understanding the culture can prove to be illuminating to understand the ecosystem, but it hardly is a factor one can directly influence. However, we can highlight technical aspects, identify axes along which common situations could be improved. In the last section, we will see how to actually cope with them.
Software culture: the foundations
As we saw earlier, software people in life science are often wet lab "defectors". Driven by domain knowledge, a backend developper is commonly more recognized by her peers when she can explain the intricacy of methylation X or Y in the chromatin rather than having deeper arguments on a sound decoupled software architecture. Well, that's a side effect of life science being ruled by life scientists (which is, for plenty of other reasons, a very good thing).
In consequence, programming language adoption, frameworks, libraries, methodology, testing, tooling and continuous building can be very far away from the state of art.
Software culture: new technologies
Hadoop, reactive programming, actors, modern web frontend, DevOps, cloud... These are not only hype words (again!). All these technologies could have been invented for life science purposes, as they perfectly answer many practical problems.
However, their adoption level is way below their promises. This is partly due to reasons described in the previous paragraphs, but we can also admit that these solutions are scary. To make the matter worse, various implementations are continuously appearing and fading away, sometimes entangled in each others. As a matter of fact, these technologies are looked at with fear and envy, like strange, dangerous but promising new worlds.
Without engaging in a long discussion on the topic, we can still make one last observation. The fantastic innovation level in the lab methods, the imagination demonstrated when designing a new drug are far more disruptive than changing an IT technology. Why don't we find in the IT system of life science the creativity and disruptiveness one handles when creating drugs?
Software development methodology
"Agile" is like "big data": yet another buzzword. But there are reasons why the word took such a momentum. Projects rarely fail only because of the lack of developers' low level technical skills [21]. It's the overall organization of the project, and even the overall organization of the information system at the institution scale, that often carries in its genes the reasons of a digital project success (or failure).
We will not try to describe here the agile philosophy and its countless implementations (a massive literature does exist, indeed), as the Agile Manifesto [22] gives the best possible introduction. Every aspect of the agile approach makes it perfectly suitable for building digital projects in the life science field.
However, "Agile" is not just a word and pronouncing it like an incantation in a yearly meeting is not enough to transform a whole corporate culture [23]. Among the most frequent failure patterns, we can observe:
- a lack of top-down support: a development team tries to adopt an Agile methodology but lacks support to open smooth communication channels with stakeholders and customers . The management does not embrace the "respond to change" nor the "customer collaboration" paradigms and feels uncomfortable without long term requirements set in stone and a matching well defined budget.
- Only top-down pressure: Agile is presented as a new religion by the management and the way to exorcise all old evils. But no room is made for training people and transforming the organization. Two years later, the converted manager flies to another position, the then out-of-fashion religion is banished and the team moves back to the old ways.
So what. Now what?
Addressing the technical aspects
Software crafting
The software skills can be addressed at the individual level. Many tools do exist: organizing a biweekly forum, pairing developers to cross-pollinate know-how, reviewing code to increase quality, arranging 20% of blue sky time slots or bringing an outside coach for specific needs.
Challenging established software practice is an investment. It will slow down productivity on the short term, but the return is soon to be high on software quality, productivity and employee motivation. Last but not least, seeking technical excellence is certainly the most efficient way to hire the best talents... and to keep them.
Our experience shows that evaluating a new technology is even harder to pursue in house than improving one's specific skill. More than a fearless and motivated lone software engineer, it relies on actual experience with the technology, field comparisons with alternatives and a clear picture of how it would fit in a global architecture. Outside help can therefore be a solution to consider before making long term radical commitments.
Methodology: moving to Agile
No out-of-the-box method exists and technical processes shall be adapted to the people, the structure, the company business and culture. But even if software development cycles are paced by bi-weekly sprints, if team members stand up every morning for a ten minutes chat and if a software factory continuously deliver working software, it does not mean that the Agile transition is over.
Agile is a philosophy, a mindset ingrained at every level in a digital company. Roles are often to be redefined, processes redesigned, communication channels widened and transparency embraced. Not only top management must be committed to it, but all the concerned actors must understand and be part of it.
It does not mean that the whole organization has to change at once. A pilot project can be launched to test and seed the approach. However, within this project boundaries, from the top management sponsor to the software engineers and in house customers, roles and relationships have to be adapted.
We deeply believe in the benefits of such a transformation. It has proven to be efficient in many domains and organizations and fits particularly well the challenges at stake in life science. But we also have seen how such a transformation is multi dimensional. Therefore, seeking outside guidance might be an option to consider.
Life Science's digital situation is not unique
At this stage, you should be convinced that life science challenges are unique... only to some extent. Our day-to-day experience with clients in other industry is actually very similar. If their business is different (and often much simpler than biology), banks, retailers, insurances, social media companies are faced with the same issues and challenges.
They also often struggle to make their digital organisation evolve. Yet their situation is strikingly similar, as the ultimate goal is to keep pace with customer evolving needs, in a sustainable way. Therefore, they also need to:
- build modern, interactive and useful tools;
- handle massive flow of complex data;
- have transparent knowledge shared across stakeholders;
- continuously deliver running software;
- and not forget to have some fun.
[caption id="attachment_56455" align="aligncenter" width="400"] 
Towards digitalisation.
[/caption]
Boarding for a new journey?
The journey towards digitization is a great and perilous one but the software industry has matured and the time has come to embrace it. By the complexity of the underlying scientific domain, the data heterogeneity, its volume and analysis processes, the life science industry would benefit greatly from all those new trends and architectures. With a careful and educated strategy, some companies have demonstrated the benefit of such an endeavor.
Are you ready to embark?
References
- [1] "The Importance of the Pharmaceutical Industry for Switzerland", S. Vaterlaus et al, 2011
- [2] Swiss Biotech Report 2015 http://www.swissbiotechreport.ch
- [3] the citation is repeated as a meme, but we could not locate the original reference
- [4] http://www.campusbiotech.ch/en/
- [5] http://epfl-innovationpark.ch/
- [6] http://www.biopole.ch/en/index.html
- [7] http://www.forumdes100.com/2010/03/health-valley-romande-la-carte.html
- [8] Health2020, Federal Office of Public Health http://www.bag.admin.ch/gesundheit2020/index.html?lang=en
- [9] https://www.humanbrainproject.eu/
- [10] http://www.chuv.ch/biobanque
- [11] "Au CHUV, une nouvelle biobanque unique en Europe", Le Temps, 12/13/2012
- [12] "The $1,000 genome", E.C. Hayden, Nature 3/19/2014
- [13] Allen Brain Atlas http://www.brain-map.org/
- [14] "Best science graphics visualization" Wired, 2014
- [15] Visualizing Biological Data conference http://vizbi.org/
- [16] "Le dossier électronique du patient arrive" J.Y. Gabbud, le Nouvelliste, 8/27/2015
- [17] https://www.partipirate.ch/2015/08/30/nouveau-dossier-medical-eletronique-valaisan-la-protection-du-patient-bradee/
- [18] "Esther Waeber-Kalbermatten suspend le projet cyber-santé en Valais" M. Atmani, Le Temps, 8/31/2015
- [19] https://en.wikipedia.org/wiki/Quantified_Self
- [20] "What Differences Make a Difference?" E. Mannix & M.A Neale, Psychological Science In The Public Interest, 2005
- [21] "The Most Common Reasons Why Software Projects Fail", T. Putnam-Majarian & D. Putnam, InfoQ, 6/13/2015
- [22] http://www.agilemanifesto.org/
- [23] "8 Reasons Why Agile Projects Fail" L. Cunningham, 4/9/2015 http://blogs.versionone.com/agile_management/2015/04/09/8-reasons-why-agile-projects-fail/