Geo localizing Medline citations
Where are the scientific publications coming from? Geolocalizing Medline citations

Big Data is not only a buzz word. A rich ecosystem of tools have emerged, together with new architectural paradigms, to tackle large problems. Open data are flowing around, waiting for new analysis angles. We have focused on the Medline challenge to demonstrate what can be achieved.
To provide some insights on how an interactive web application was built to explore such data, we will discuss the geographic localization method based on free text affiliation, Hadoop oriented treatment with Scala and Spark, interactive analysis with the Zeppelin notebook and rendering with React, a modern JavaScript framework. The code has been open sourced on github [1, 2] and the application is available on Amazon AWS.
Introduction
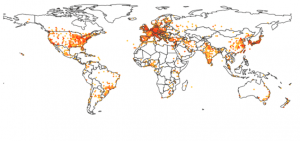
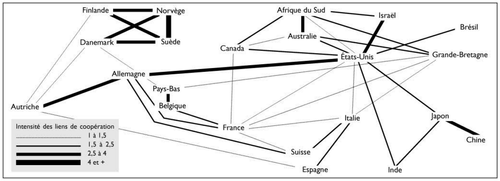
Analyzing the geographic origin of scientific publications has been addressed in previous papers, such as [3, 4, 5]. Although these papers present an interesting sociological, political and historical view on the matter, the underlying data has been aggregated by hand (see figure 1) and offers little possibilities of further digging.
More comprehensive dataset are nonetheless available, such as arxiv.org [6], but we decided to focus on the biology and biomedical citation repository Medline. As the underlying source of data for Pubmed [7], the major entry point for any bibliographic research in the field, it offers both a large volume of citations and a parsable format.
If countless questions can be digged out of such materials, we were wondering which countries are the most prolific across the years. Furthermore, are there any patterns in country collaborations? Even though localization information is not directly made available, it can be deduced from the authors’ affiliations in a majority of cases (when available). Finally, we wanted to provide both an interactive tool to navigate through those data and a framework to analyze them in a programmatic way.
The current article has no other claims than offering a visualization tool and an access to that data for further analyses.
Materials
Citations data were downloaded from Medline archives. They can be made available via an NLM agreement [8]. Mirrored in October 2015 for this study, they consist in 789 files (15GB) containing 23 millions citations in XML format. Each citation is comprehensive, does not include the article content but meta information such as publication date, title, authors and partial affiliation, references etc. An example of such an output can be found here [9].
Localizing the publication with geographical coordinates and country is derived from the author’s affiliation, whenever available. Details on this process will be discussed below, but two major sources of localization were used. If the Google map geocoding API [10] is convenient for this purpose, limitations on the free usage forbids a large scale usage. A first home made geocoding pass was therefore designed to parse the free text affiliation fields, based on GeoNames dumps [11].
The last source of external data used in this project are country geographical shapes, derived from the Natural Earth Data repository [12].
Finally, such a programming project relies on countless bricks of open source software. Without citing them in details, the main components are Play [18], Spark [16] and the Scala language for the backend. The front end web application was developed in JavaScript, using the React library[13] and heavily relying on d3.js [14] for graphical components. Although the published code was executed on a laptop (with Solid State Disk), the used technology is originally aimed to connect to an Hadoop cluster, for the sake of scalability.
Data crunching
The data crunching step consists in downloading original data and turning them into an accessible and exploitable format. In this article, it mainly consists in importing citations and assigning one or more geographical locations whenever possible.
Method
Download Medline archives
As mentioned in the material section, Medline citation archive can be downloaded (once the license agreement has been exchanged). Although the method seems trivial, the command for the sake of easier reproducibility is:
lftp -e 'o ftp://ftp.nlm.nih.gov/nlmdata/.medleasebaseline/gz && mirror --verbose && quit'
Geo localization
One key step of data preparation is to assign a latitude, a longitude and a country to citations. Medline citations XML format exposes a list of authors, such as for [9]. One of the author’s details looks like:
<Author ValidYN="Y"> <LastName>Lill</LastName> <ForeName>Jennie R</ForeName> <Initials>JR</Initials> <AffiliationInfo> <Affiliation>Genentech, South San Francisco, California 94080, USA.</Affiliation> </AffiliationInfo> </Author>
The Affiliation field is therefore to be mapped to geographical coordinates.
Google GeoCoding API
The easiest way to translate an address text field into such coordinates is to use Google Geocoding API [10]. It is the underlying layer mapping an address to a position when using the Google map or Google Earth applications.
A REST API is available, accessible via a URL like:
https://maps.googleapis.com/maps/api/geocode/json?address=Genentech,+South+San+Francisco,+California+94080,+USA&key=YOUR_API_KEY
Where the YOUR_API_KEY value is to be requested by registering to the Google API system. The JSON output can be parsed for resolution (is the address unequivocally resolved? which coordinates? which country?). A Java API is also available to leverage the parsing and make the process even simpler [15].
However, a big limitation is the Google free plan over the API. The daily limit is defined to be 2’500 requests, hardly usable when approximately eight millions unique affiliation addresses are to be resolved. Other online solutions exists (such as MapQuest) but suffers from the same usage limitations or were shown to be too strongly biased towards the United States. A handmade offline solution had therefore to be designed.
Our implemented geocoding method relied on
- the attempt to resolve affiliations by the GeoNames solution described in the next section;
- the unresolved affiliations were then sorted by recurrence (the most common first) and a daily call to the Google API made to try to locate the next 2500 top most. For the sake of this project and due to time constraints, this second step has only been applied for a few days, to test the approach relevance.
A GeoNames based solution
GeoNames is a comprehensive geographical database [11]. Similarly to Google Maps API, it provides a REST API to be queried, but with similar limitations. However, the underlying database can be freely downloaded in tabular format, with several views. The ad hoc solution presented below is not perfect but proved to be relevant enough to solve most of the situations at hand in the current project.
A lot of information is available in GeoNames database (like currency, postal code format…), but only a subset of it is actually used in the current implementation. Our solution uses three files, all cross linked with proprietary id fields:
- countryInfo.txt: country ISO codes and names;
- cities15000.txt: names, latitudes, longitudes, countries and population for cities larger than 15’000 inhabitants;
- alternateNames.txt: countries and cities alternative names or spellings.
The actual algorithm to localize address field is not straight forwards for two major reasons. It is filled as a free text by authors at paper submission time, without clear indications on the format. Although they can be unequivocal for a researcher reading the paper, they offer a challenge from an automation point of view. Here are a few examples:
- “College of Physicians and Surgeons, Columbia University, New York, NY, USA”,
- “CJ. N. Adam Memorial Hospital, Perrysburg, N. Y.", without any country;
- “Tripoli”, besides various spelling, is a major city in three countries;
- email information sometimes ends up in the affiliation field;
- multiple affiliations for one author can be filed;
- Italy capital can be spelled “Rome” or “Roma”...
Many places have various common names: “New York”, “New York City”, “NYC”. “Springfield, USA” can point to eleven different towns with more than 15’000 inhabitants.
To make the problem even more intricate, some affiliations do not provide any country such as “Oxford College” or “Department of Medicine, University of Washington, Seattle 98195" or provide different depths, such as street address, postal code etc. Finally, to give an ultimate flavor of the problem, we could also mention country that do not exist anymore or had their named changed: “USSR”, “Hong Kong” or “Tchecoslovaquia”.
If the Google Maps API has proven to be efficient to disambiguate many cases, it relies on much more information, large code base and unpublished methods. Therefore, we have designed a multi steps strategy to address the most common situations. The goal is to extract city/country pairs and look out in the database for unique such matches. The major steps are:
- remove emails, head reference numbers, and extract the first affiliation when multiple are present, splitting on dots and semi columns (but no dots following initials or common abbreviations);
- split the string in blocks along comma separators;
- remove blocks with postal code structure;
- with all combinations of two elements among the last three blocks, look up in the GeoNames database for matching:
- city/country pair,
- city/country pair, with alternate names,
- city only,
- city only, with alternate names;
- the process is stopped at the first match on the previous step;
- if multiple locations are matching, they are sorted by population count and the first one is kept when the population ratio with the second is greater than 10.
As discussed below, a lot of affiliations are not resolved unambiguously, but a majority are. Imperfect as it may be, the presented method relies on empirical tradeoffs. To illustrate the method, examples can be found in the project unit tests [1].
Implementation
Out of the original Medline citations, only a few informations were kept for the sake of this project: publication date, title, abstract and author list. They were saved in parquet format, in a Hadoop hdfs fashion to allow for distributed processing with the versatile Spark framework [16]. Although it relies on hdfs oriented format, the crunching part was actually done at once on a mac laptop, executing steps located in the ch/fram/medlineGeo/crunching/tools directory [1].
Results
It is now time to review numbers and measure the enrichment of the crunched data. As the data are processed on the Hadoop platform, Apache Zeppelin was used for the analysis [17]. It is a web based notebook, allowing users to interactively manipulate hadoop hosted data while programming in Scala (Java or Python) and Spark [16].
Geo localization method performance
Out of the 23 millions citations, 8.7 millions unique affiliation fields were extracted. For example, “Department of Biology, Massachusetts Institute of Technology, Cambridge 02139” refers to 724 publications. How many of them could be attributed to a geographical location by each of the previously described methods?
65% were resolved by the custom GeoNames based strategy. Among the remaining ones, 75% were assigned by the Google Geocoding API. Although this second step has not been applied to the whole set for time reasons (it would have taken 1218 days with the free access limit!), one can predict that 91% of the overall addresses could be resolved.
We can now measure how many publications could be linked to a geographical location.
Evolutions of located citation counts through time
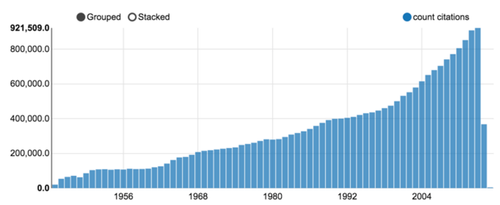
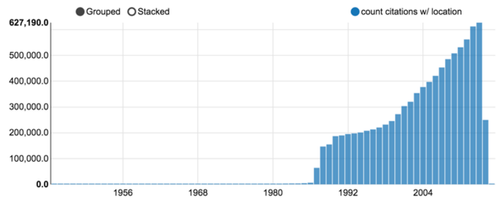
As of October 2015, more than 23 millions citations were downloaded. Although some publications from the late XIXth century were registered, the volume drastically increased after WWII, as shown in figure 3.
Figure 3: Medline citation count per year (a Zeppelin notebook chart output).
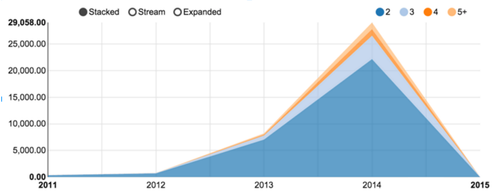
Among those 23 millions citations, 9 millions could be associated with at least one location, as shown in figure 4. Taking up in the mid eighties, the percentage is stable, around 70% of the overall production. Figure 5 shows the numbers of publications associated with two or more countries . The comparatively low numbers are due to the tendency to fill the affiliation only for the first author. Moreover, the rate of localization resolution deeply affects citation with authors coming from multiple research centers.
_ 
Browsing through geographically located publications
Once the data crunching steps are executed, citations are annotated with location and stored in parquet format. Although they can be read by several tools from the Hadoop stack, we present two components built for this project:
- a backend server: reading, transforming and serving the data through a REST API;
- a frontend rich web application, to display and interact with graphical representations, which communicate with the backend via the API.
Such a modularized approach allows to change the presentation layer, which could easily be another web application, R scripts or countless other solutions.
The backend server [1]
The backend goal is to provide access to data through an HTTP REST API. For example http://localhost:9000/countries/citationCount/2012 is expected to return a list of countries with the number of assigned publications in 2012. The output is to be structured (json) or tabular (tsv).
Another example, to extract all country collaborations for year 2002, one can head to http://localhost:9000/countryPairs/citationCount/2002 and get:
[ { "year": 2002, "nbPubmedIds": 1, "countryFrom": "DE", "countryTo": "RU", "nbPubmedIdTotalFrom": 18100, "nbPubmedIdTotalTo": 3573 }, … ]
Among many possible choices (nodejs/express, java/Spring, ruby/rails, python/django etc.), we selected the Scala/Play/Spark combination. Based on the JVM and combining both functional and object oriented paradigms, we believe Scala [20] to be a language of choice for scientific computations and more broadly, distributed programming.
Play is a web framework, for both Scala and Java. Besides practical developper comfort, it is naturally designed to serve reactive programming [20], with large, interactive and resilient transactions.
The third component of the trio is Spark [16], a powerful engine sitting originally on top of Hadoop. It allows to distribute computations over large volumes of data and is accessible in Java and Python, but the natural language is… Scala. The Spark project has got a strong traction in the recent years, leveraging Hadoop MapReduce batch processing into a more interactive and comfortable abstraction. The framework continuously grows, including event streaming, machine learning, graph algorithms and an R interface.
Aggregating data via Spark launches a Hadoop job. Even though the framework made huge speed improvements over the original MapReduce system, it is hardly enough for a convenient interactive usage (common response times can be as long as a minute). Therefore, all classic views are computed once, cached and can then be served within a tenth of a second.
As mentioned a couple of times, the selected storage architecture for this project was Hadoop oriented. It shall nonetheless be mentioned that this choice could be challenged. The volume at hand is not that large and other solutions, such as ElasticSearch [21] could be considered. We gave an attempt to mongodb [22] but data aggregation did not show to scale up.
The frontend web browser [2]
The second component consists of a rich web application. Once again, modern Javascript offers a plethora of frameworks, such as AngularJS, Ember or Backbone. Although all of them can show clear advantages in some situations, we opted for React [13], an open source rendering library provided by Facebook. Together with the Flux design pattern [23], it has proven to meet the requirements of massive data display. In complement, one can hardly avoid the versatile d3.js library [7] to build interactive graphical representations.
We propose three different type of views:
- a world map, with a heat map to represent citation counts (figure 6);
- a bar chart, with publication counts per year (figure 7);
- a network graph, displaying collaborations among countries (figure 8).
Each of these views is enriched by a year selection slider to navigate in the past and mouse interactions to reveal more details.
A world heat map
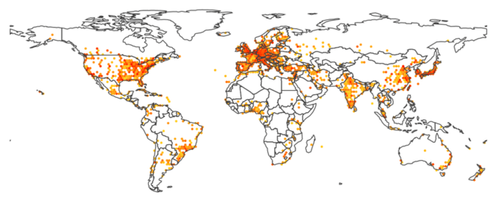
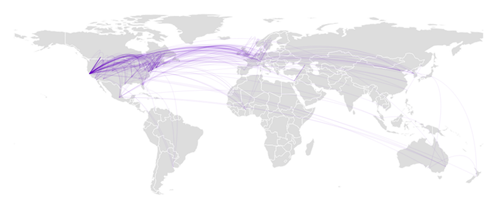
Figure 6: a world heat map, where colors ranging for light orange to red show the number of publications for the current year.
Once citations could be assigned to geographical coordinates, it felt natural to display them on a world map. The results presented in figure 6 shows an hexagonal paving, with the color indicating the local number of publications in the area. Using the slider to navigate across years reveals for example the recent dramatic increase of production from the BRICS countries.
D3.js is combined with topojson for a near optimal rendering of country borders, allowing to superimpose other information, such as the links displayed in figure 2.
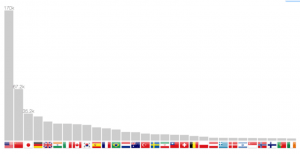
Publication count per country
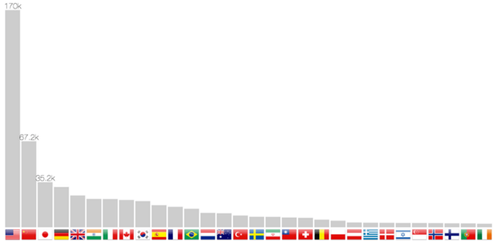
The localization method allows not only to return coordinates, but also the country. It is therefore possible to build a bar chart in figure 7. Scrolling through the years makes the bars slide from one position to another, therefore following one country progression, such as China in the last twenty years.
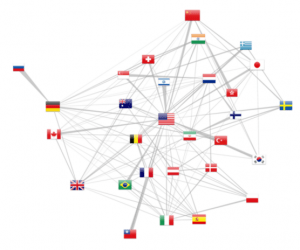
Country collaboration graph
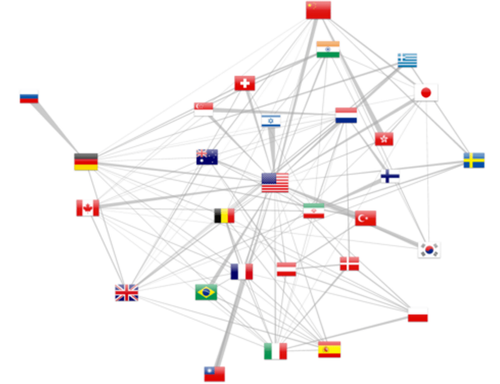
Finally, multiple affiliated citations allow to pull out cross country collaborations (at least as of 2012, as revealed by figure 5). The force layout attracts more strongly countries with more collaborations while the overall number of publications is reported by the flag size. Moreover, d3.js allows to draw trapezoidal edges, where the thickness of one end shows the relative importance of the collaboration. For example, in the upper left corner, Russia is exclusively linked with Germany, whereas this latter country as other collaborations. The same observation can be made for the Israel-USA relation.
One should keep in mind that this representation is only a sample of the actual human co-publishing, as either the authors might not have filed multiple affiliations or they could have not been solved by the localization method. We can only only address a sample of the actual collaborations. Numbers are nonetheless more relevant in the recent years, following the increase of citations with multiple affiliated authors. Finally, the 65% address elucidation rate for a single address falls down to 42% for publications with two addresses (0.652). Extending the geo localization method to Google Map API would therefore further resolved 83% such cases.
Running the application
For development purpose, both components can be cloned out from github [1, 2] and started independently, as described in their respective README.md files.
For a more convenient usage, a docker image can be pulled out and ran from http://hub.docker.com. This image has been published for demonstration only. To limit memory and CU footprint, the JVM backend is not packed and data are cached within the NodeJS application:
docker run -p 80:5000 alexmass/medline-geo-standalone
Continuous testing is hosted by Travis [24], an online continuous integration system with free access for open source projects.
Conclusions
We have presented an approach to attribute geographical information to scientific citations, based on the Medline citation index. Moreover, we have seen how a Big Data stack can be used with a modularized architecture via an API data exposition.
If those data can reveal interesting phenomena, we believe that this approach can be extended to dig further on this data set or others (arxiv.org), analyzing abstract contents for example, or aggregating other sources of information (such as yearly GDP).
At a larger scale, we have demonstrated how the Hadoop ecosystem paves new routes to dive into the analysis of massive datasets, at a relatively low cost.
Acknowledgments
I’d like to thank many contributors for the presented work. Among them, Nasri Nahas and Catherine Zwahlen for seeding the idea. Martial Sankar for enlightening discussions concerning the geo localization methods. Last but not least, my colleagues at OCTO Technology Switzerland for their thorough feedback.
References
[1] https://github.com/alexmasselot/medlineGeoBackend [2] https://github.com/alexmasselot/medlineGeoWebFrontend [3] “Les formes spécifiques de l’internationalité du champ scientifique”, Yves Gingras. Actes de la recherche en sciences sociales, 2002 [4] “L’internationalisation de la recherche en sciences sociales et humaines en Europe (1980-2006)”. Yves Gingras et Johan Heilbron. L’espace intellectuel en Europe, 2009 [5] “International scientific cooperation : the continentalization of science”, M. Leclerc et J. Gagné. Scientometrics, vol. 31, 1994, p. 261-292. [6] arXiv.org http://arxiv.org/ [7] PubMed http://www.ncbi.nlm.nih.gov/pubmed [8] https://www.nlm.nih.gov/bsd/licensee/access/medline_pubmed.html [9] http://www.ncbi.nlm.nih.gov/pubmed/25428506?report=xml&format=text [10] https://developers.google.com/maps/documentation/geocoding/intro [11] http://www.geonames.org/ [12] http://www.naturalearthdata.com/ [13] https://facebook.github.io/react [14] http://d3js.org/ [15] https://github.com/googlemaps/google-maps-services-java [16] http://spark.apache.org/ [17] https://zeppelin.incubator.apache.org/ [18] https://www.playframework.com/ [19] http://www.scala-lang.org/ [20] http://www.reactivemanifesto.org/ [21] https://www.elastic.co/ [22] https://www.mongodb.org/ [23] https://facebook.github.io/flux/docs/overview.html [24] http://travis-ci.org